
Tags: Homology | Fungi | Sequence analysis | Biological networks
ggMatch finds reciprocal best blast/diamond hits across a large number of genomes, in an iterative fashion. The iterative nature removes the need to do the dreaded all-vs-all blast between all genomes.
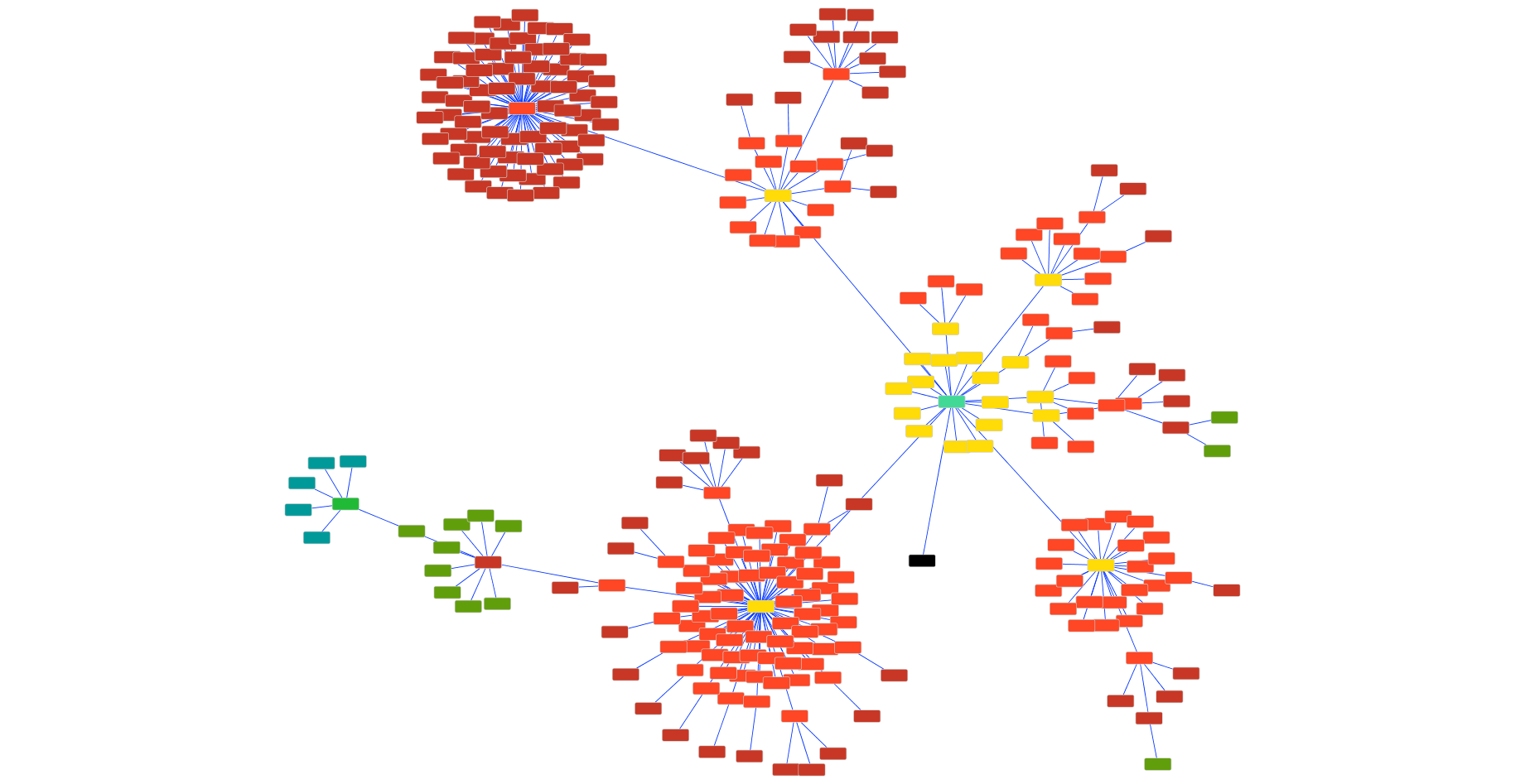
The figure below shows an example gene matching process for a single gene against 298 fungal genomes extracted from the JGI. The black node represents the original query sequence, and edges between other nodes represent high quality reciprocal best blast hits. Each node color represents a different iteration. In the first iteration, we discover the yellow genes, and in later iterations we discover other genes based on the newly discovered genes. A traditional reciprocal best hit search would have revealed only the yellow nodes in this network.